Research Interests and Results
Research interests
The laboratory is focusing attention on the biodiversity of lichens, systematic and evolutionary biology of lichen-forming fungi, desert lichens and their sand-fixating biology, and the biological activities of lichen secondary metabolites.
The following research programs are being carried out: the systematic and evolutionary biology of worldwide Umbilicariaceae lichens; molecular evolutionary biology of lichen-forming and non lichen-forming fungi; Flora Lichens Sinicarum; isolation and culture of mycobionts and photobionts from desert lichens; screening and clone of the genes in response to adversities; and the study of activity compounds from lichens, mycobionts or photobionts.
Research Results
A new genus Cetradonia was published based on the combination of phenotype and genotype, and the genus Cetradonia was placed in Cladoniaceae.
Umbilicariaceae was isolated from Lecanorales based on the combination of phenotype and genotype, and a new order Umbilicariales was described.
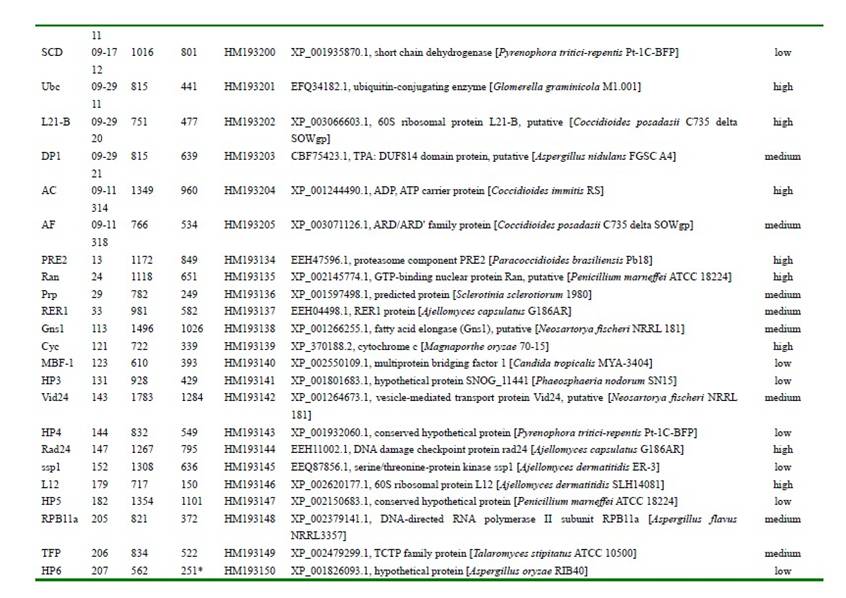
The photobiont from Endocarpon pusillum which is dominant in desert was proved to be Diplosphaeria chodatii. The isolated mycobiont form E. pussium can survive for 7 months under the stress conditions in combination with both the desiccation and starvation. The first full-length cDNA library for lichenized fungi was constructed from cultured mycobiont of the arid desert lichen Endocarpon pusillum, and 111 genes of the lichenized fungi were identified for the first time, among which 11 genes shared no homology with any known fungal genes.
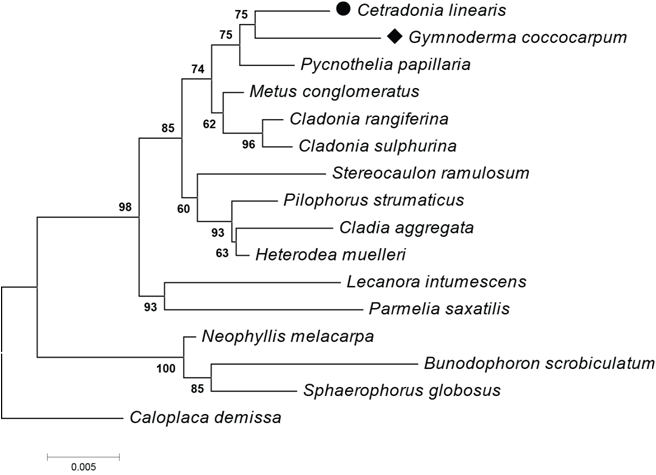
Fig.1. Minimum Evolution tree based on SSU rDNA sequence data. Bootstrap support values are shown at nodes.

Fig.2. The NJ tree inferred from SSU rDNA data. The reliability of the inferred tree was tested tree was tested by 1000 bootstrap replications; the numbers in each node represent bootstrap support values. Only bootstrap values greater than 50% are shown.
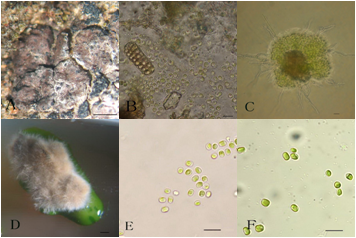
Figure 3 The lichen E. pusillum and its symbionts. A, The habit of E. pusillum; B, The ascospores with hymenial algal cells in a perithecium; C, Germinating ascospores with numerous algal cells ejected from the perithecium; D, A colony formed from the discharged ascospores together with the hymenial algal cells for 2 months after being transferred; E, The algal cells from E. pusillum in the liquid BBM; F, The algal cells of D. chodatii (UTEX 1177) in the liquid BBM. Scales: A and D = 1mm; B, C, E and F = 10µm.
Table 1. Genes in cDNA clones of Endocarpon pusillum based on their CDS comparison with other fungal genes through BLASTP search against the protein databases at NCBI.




Honours and awards



