
Research Interests
1. The genetic diversity and adaptive evolution of budding yeast, and the molecular mechanisms of microbial co-evolution.
2. The mechanisms of epigenetic inheritance and cellular identity and fate determination.
Education/degrees
2012.09 – 2018.06, Ph.D. in Microbiology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
2008.09 – 2012.06, BS in Bioengineering, School of Biological Science and Engineering, South China University of Technology, Guangzhou, China
Work experience
2024.02 – Present, Principal Investigator, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
2019.09 – 2024.02, Postdoctoral Fellow, Institute for Cancer Genetics, Columbia University, New York, USA
2018.07 – 2019.09, Assistant Researcher, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
The main research areas
1. The genetic diversity and adaptive evolution of budding yeast, and the molecular mechanisms of microbial co-evolution.
The wild and domesticated populations of budding yeast (Saccharomyces cerevisiae) have been extensively studied in terms of ecology, population genetics, and population genomics. However, there is still controversy in the academic community about the origins of domesticated populations. Our group plans to utilize NGS and third-generation sequencing technologies to obtain genomic information. Our aim is to study the basic patterns of genetic diversity and adaptive evolution in budding yeast, as well as the conservation and differences in the evolutionary patterns between species; and to elucidate the molecular mechanisms of multispecies co-evolution, especially variations in genomic structure and gene expression regulation patterns.
2. The mechanisms of epigenetic inheritance and cellular identity and fate determination.
Epigenetic information is mainly stored in chromatin through chemical modifications, such as DNA methylation and histone modifications. In budding yeast, epigenetic information is primarily transmitted and carried by parental histones to ensure the inheritance of gene expression, the stability of chromatin structure, and the maintenance of cellular characteristics. Although two main parental histone transmission pathways (Mcm2-Ctf4-Polα and Pol ε pathway) have been revealed in both budding yeast and mammalian cells, the molecular mechanisms of how parental histones are accurately transferred to the replicated DNA strands and how the modifications on the parental H3-H4 dimers are precisely duplicated onto the new (H3-H4)2 tetramers remain unsolved mysteries. Currently, the epigenetic regulation and population genetic structures in budding yeast are mostly concentrated on systematic independent research, with very few studies investigating the correlation between these two factors, hence there is a very limited understanding of their interactions. Our team intends to examine the regulatory mechanisms underlying epigenetic inheritance and to contrast the contributions of epigenetic and genetic variations in the evolutionary processes of yeast.
Representative Publications (#co-first author)
1. Duan SF, Han PJ, Wang QM, Liu WQ, Shi JY, Li K, Zhang XL, Bai FY*. The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nature Communications. 2018, 9(1):2690.
2. Duan SF, Shi JY, Yin Q, Zhang RP, Han PJ, Wang QM, Bai FY*. Reverse evolution of a classic gene network in yeast offers a competitive advantage. Current Biology. 2019, 29(7):1126-1136.e5.
3. Serra-Cardona A, Duan SF#, Yu CH, Zhang ZG*. H3K4me3 recognition by the COMPASS complex facilitates the restoration of this histone mark following DNA replication. Science Advances. 2022, 8(18):eabm6246.
4. Xu XW, Duan SF#, Hua X, Li ZM, He R, Zhang ZG*. Stable inheritance of H3.3-containing nucleosomes during mitotic cell divisions. Nature Communications. 2022, 13(1):3533.
5. Mo Y, Duan SF#, Zhang X, Hua X, Zhou H, Wei HJ, Watanabe J, McQuillan N, Su ZY, Gu W, Wu CC, Vakoc CR, Hashizume R, Chang K, Zhang ZG*. Epigenome programming by H3.3K27M mutation creates a dependence of pediatric glioma on SMARCA4. Cancer Discovery. 2022, 12(12):2906-29.
6. Yang X, Duan SF#, Li ZM, Wang Z, Kon N, Zhang ZG*, Gu W*. Protocol of CRISPR-Cas9 knockout screens for identifying ferroptosis regulators. STAR Protocols. 2023, 4(4):102762.
7. Li Z, Duan SF#, Hua X, Xu XW, Li YL, Menolfi D, Zhou H, Lu C, Zha S, Goff SP, Zhang ZG*. Asymmetric distribution of H3K9me3 at replicating DNA strands silences L1 retrotransposons during S phase. Nature. 2023, 623:643-651.
Other collaborative publications
1. Han DY, Han PJ, Rumbold K, Koricha AD, Duan SF, Song L, Shi JY, Li K, Wang QM, Bai FY*. Adaptive gene content and allele distribution variations in the wild and domesticated populations of Saccharomyces cerevisiae. Frontiers in Microbiology. 2021, 12:631250.
2. Song L, Shi JY, Duan SF, Han DY, Li K, Zhang RP, He PY, Han PJ, Wang QM, Bai FY*. Improved redox homeostasis owing to the up-regulation of one-carbon metabolism and related pathways is crucial for yeast heterosis at high temperature. Genome Research. 2021, 31(4):622-634.
3. Yang X, Wang X, Li Z, Duan SF, Li H, Jin J, Zhang Z, Gu W*. An unexpected role for Dicer as a reader of the unacetylated DNA binding domain of p53 in transcriptional regulation. Science Advances. 2021, 7(44):eabi6684.
4. Evrin C, Serra-Cardona A, Duan SF, Mukherjee PP, Zhang Z*, Labib KPM*. Spt5 histone binding activity preserves chromatin during transcription by RNA polymerase II. EMBO Journal. 2022, 41(5):e109783.
5. Bai FY*, Han DY, Duan SF, Wang QM. The ecology and evolution of the baker’s yeast Saccharomyces cerevisiae. Genes. 2022, 13(2):230.
6. He PY, Shao XQ, Duan SF, Han DY, Li, K, Shi JY, Zhang RP, Han PJ, Wang QM, and Bai FY*. Highly diverged lineages of Saccharomyces paradoxus in temperate to subtropical climate zones in China. Yeast. 2022, 39, 69-82.
7. Su ZY, Kon N, Yi JJ, Zhao HQ, Zhang WW, Tang QS, Li H, Kobayashi H, Li ZM, Duan SF, Liu YQ, Olive KP, Zhang ZG, Honig B, Manfredi JJ, Rustgi AK, Gu W*. Specific regulation of BACH1 by the hotspot mutant p53(R175H) reveals a distinct gain-of-function mechanism. Nature Cancer. 2023, 4:564-581.
8. Yang X, Wang Z, Zandkarimi F, Liu YQ, Duan SF, Li ZM, Kon N, Zhang ZG, Jiang XJ, Stockwell BR, Gu W*. Regulation of VKORC1L1 is critical for p53-mediated tumor suppression through vitamin K metabolism. Cell Metabolism. 2023,35(8):1474-1490.e8.
9. Yang X, Wang Z, Samovich SN, Kapralov AA, Amoscato AA, Tyurin VA, Dar HH, Li ZM, Duan SF, Kon N, Chen D, Tycko B, Zhang ZG, Jiang XJ, Bayir H, Stockwell BR, Kagan VE, Gu W*. PHLDA2-mediated phosphatidic acid peroxidation triggers a distinct ferroptotic response during tumor suppression. Cell Metabolism.2024.
Representative Pictures
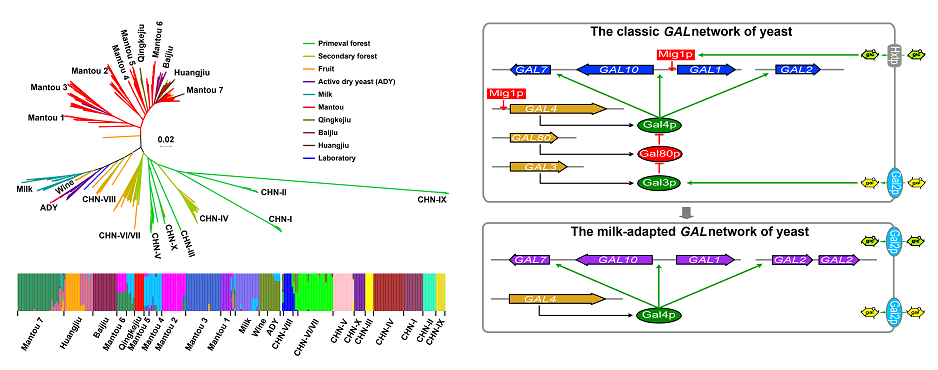
Duan et al., Nature Communications, 2018 Duan et al., Current Biology, 2019
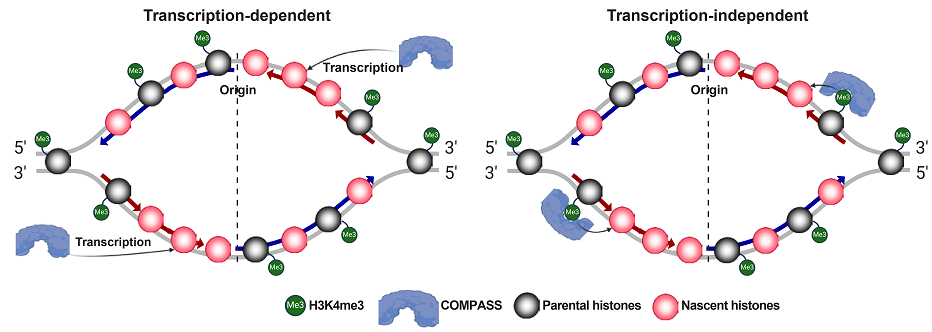
Serra-Cardona and Duan et al., Science Advances, 2022
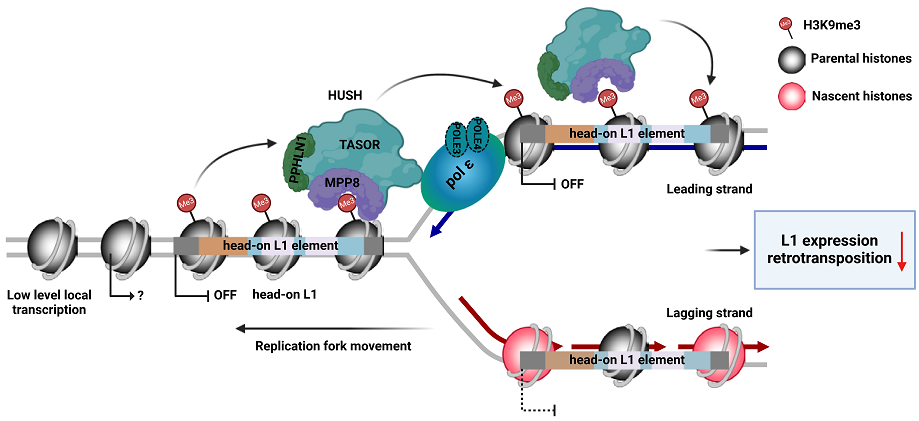
Li and Duan et al., Nature, 2023



